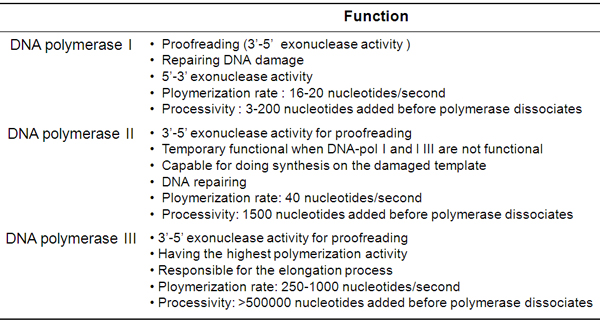
Large fragment is called Klenow fragment. It possess 5΄ --> 3' DNA polymerization activity and 3΄ --> 5΄exonuclease activity for proofreading, but 5' --> 3' exonuclease activity is lost.
Small fragment retains 5΄- --> 3΄ exonuclease activity but does not have the other two activities exhibited by the Klenow fragment (i.e. 5'- --> 3' polymerase activity, and 3- --> 5' exonuclease activity).
The Klenow fragment of DNA polymerase I is widely used in molecular biology.
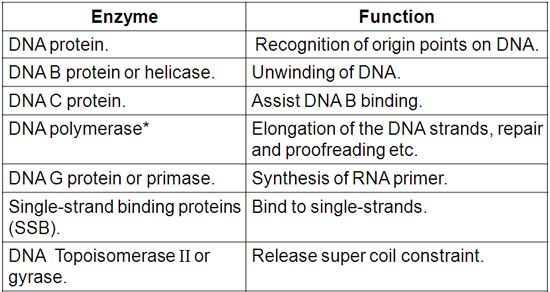
DNA-pol α: initiate replication and synthesize primers.
DNA-pol β: replication with low fidelity.
DNA-pol γ: polymerization in mitochondria.
DNA-pol δ: elongation.
DNA-pol ε: proofreading and filling gap.
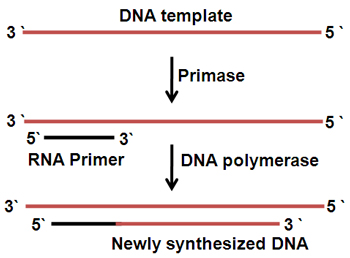
Primers are short RNA fragments of a several nucleotides long. Primase is able to synthesize primers using free NTPs as the substrate and the ssDNA as the template.
Primers provide free 3΄-OH groups to react with the α - P atom of dNTP to form phosphoester bonds.
Primase, DNAB, DNAC and an origin form a complex at the initiation phase.
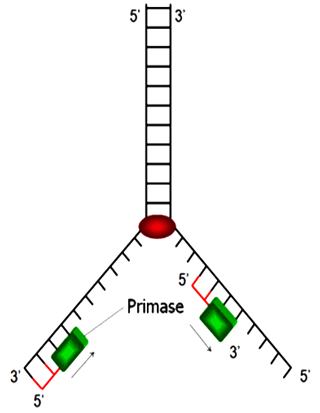
SSB protein : To avoid the recontact of the strands opened by helicase, single strand DNA binding protein attach to each single strand preventing the reformation of dsDNA. It also protects the vulnerable single stranded DNA against nucleases.
Topoisomerase II or DNA gyrase : Topoisomerase II are functional in releasing the torsional strain generated because of unwinding of the dsDNA. This also prevents the formation of supercoil ahead of replication forks.
DNA Ligase : Connects two adjacent ssDNA strands by joining the 3΄-OH of one DNA strand to the 5΄-P of another DNA strand. It also is responsible for sealing the nick in the process of replication, repairing, recombination, and splicing.